Single-cell meta-analyses reveal responses of tumor-reactive CXCL13+ T cells to immune-checkpoint blockade
2022-09-25
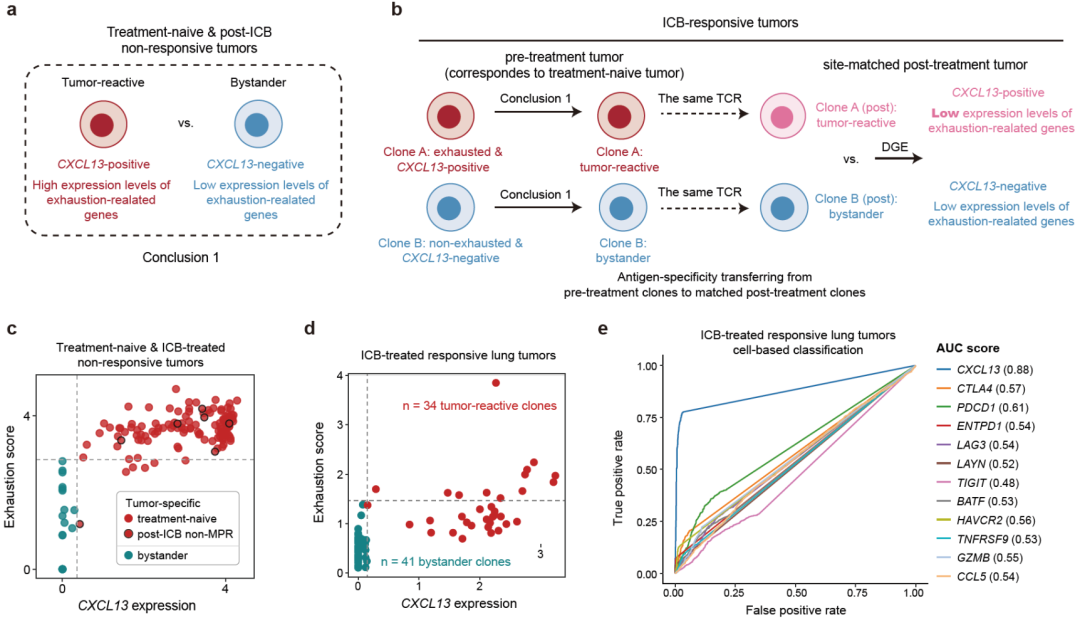
CD8+ T cells are responsible for the recognition and killing of cancer cells. Following persistent exposure to its cognate tumor antigen in cancer, a CD8+ T cell will undergo differentiation towards a dysfunctional status, which is marked by the up-regulation of co-inhibitory molecules and by the progressive loss of proliferative capacity and certain effector functions1. Immune-checkpoint blockade (ICB) is capable of targeting those negative regulatory proteins such as PD-1, thereby enhancing immune responses in multiple cancers2,3. However, the fundamental mechanisms underlying ICB remain incompletely understood, hindering the development of more effective immunotherapies.Single-cell RNA sequencing (scRNA-seq) is a versatile technique to dissect the cellular diversity of the tumor microenvironment (TME), enabling the characterization of the temporal dynamics of T cells following ICB. As single-cell studies are extended to multiple cancer types, patterns start to emerge that distinctive T cell responses to ICB may exist in different cancer types. For example, recent analyses of basal cell carcinomas (BCCs)4 and breast tumors5 revealed that anti-PD-1 predominantly induced an increase in exhausted CD8+ T (Tex) cells after effective treatment, whereas in non-small-cell lung cancer (NSCLC)6, Tex precursors were the ones that selectively respond to PD-1 blockade. In addition,the published analysis4 of the BCC data demonstrated that new tumor-specific clones appeared after treatment, with clonotypes different from those detected in pretreatment tumors. However, this clonal replacement phenomenon has not frequently been observed in lung6 and breast tumors7. It therefore becomes critical to perform meta-analyses of immunotherapy single-cell datasets to enable systematic comparison of T cell responses to ICB across multiple cancer types.While single-cell analyses yielded insights into T cell behaviors, in most cases, their antigen specificities are largely unknown, which hinders the understanding of tumor-reactive T cell responses, especially considering the prevalence of bystander T cells within tumors8. Multiple studies8–12 have been specifically conducted to characterize the phenotypes of tumor-reactive CD8+ T cells and have suggested that exhaustion-related genes, for example, CD39 (8,12), are capable of discriminating tumor-reactive from virus-specific bystander clones within treatment-naive tumors. However, within certain ICB-responsive tumors, tumor-reactive CD8+ T cells are in a ‘non-exhausted’ state due to the effect of ICB, expressing much lower levels of exhaustion-related genes6,13. Therefore, it remains a challenge to accurately identify tumor-reactive CD8+ T cells from different contexts. Here, by analyzing our data and recently published scRNA-seq datasets with the T cell antigen specificity information, we showed that measuring CXCL13 expression could effectively identify both exhausted and non-exhausted tumor-reactive CD8+ T cell clones within tumors. Using scRNA-seq data generated by us and others, we further constructed an immunotherapy single-T-cell compendium across five distinct human cancer types. Our analyses revealed temporal behaviors of CXCL13+ tumor-reactive T cells across cancers, providing insights into T cell-intrinsic mechanisms of sensitization and resistance to ICB.